(ref 10-0023)
Specifications:
- Carbapenemases*: KPC, NDM, VIM, IMP, OXA-48-like, MCR, GES, GIM, SPM, OXA-23-like, OXA-24/40-like, OXA-58-like
- CTX-M ESBLs: CTX-M-1 group, CTX-M-1-like, CTX-M-15-like, CTX-M-3-like, CTX-M-32-like, CTX-M-2 group, CTX-M-8 & -25 group, CTX-M-9 group
- TEM ESBLs vs. non-ESBL: TEM wt, TEM E104K, TEM R164S, TEM R164C, TEM R164H, TEM G238S
- SHV ESBLs vs. non-ESBL: SHV wt, SHV G238S, SHV G238A, SHV E240K
- Other ESBLs**: VEB, PER, BEL, GES
- AmpCs: CMY I/MOX, ACC, DHA, ACT/MIR, CMY II, FOX
- Controls included: DNA control, Amplification control, Hydridization control, Negative control
- Specimen: Culture
- Sample preparation: Magnetic bead- or column-based methods***
- Pre-PCR equipment: Thermocycler***, vortex mixer, mini-centrifuge
- Post-PCR equipment: Thermocycler***, vortex mixer, mini-centrifuge, thermomixer with active cooling***, Check-Points Tube Reader including E-Ads software, computer with USB drive and internet connection, barcode reader (optional)
- Throughput: 1 to 24 samples/run
* MCR-1, 2; KPC-2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 21, 22, 23, 24, 25; OXA-48, 48b, 162, 163, 181, 204, 232, 244, 245, 370; VIM-1, 2, 3, 4, 5, 6, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47; NDM-1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16; IMP- 1, 3, 6, 7, 8, 10, 13, 19, 20, 24, 30, 37, 40, 42; GES-2, 4, 5, 6, 13, 14, 15, 16, 18, 20, 21; GIM-1, SPM-1; OXA-23, 27, 49, 73, 146, 165, 166, 167, 168, 169, 170, 171, 225, 239; OXA-24, 25, 26, 33, 40, 72, 139, 207; OXA-58, 96, 164.
** VEB-1, 2, 3, 4, 5, 6, 7, 8; PER-1, 2, 3, 4, 5, 6; BEL-1, 2, 3; GES-1, 3, 7, 8, 9, 10, 11, 12, 17, 19, 22.
*** contact Check-Points for specifications: info@check-points.com
Workflow:
|
Time per step |
Hands-on time per step |
| 1. Culture |
|
|
2. DNA extraction
of total nucleic acid using automated, magnetic bead or column-based methods (not supplied) |
|
|
3. Identification
by multiplex ligation |
3 h |
15 min |
4. Amplification
of ligated probes by PCR |
1.5 h |
15 min |
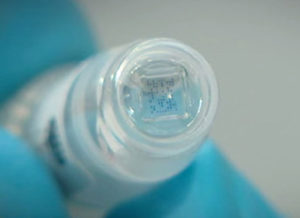
5. Detection
through hybridization of amplified probes to specific locations on the microarray, contained in a Check-Points Array Tube |
2 h |
30 min |
6. Results
are generated using the Check-Points Tube Reader to produce an image of the microarray and the E-Ads software to automatically translate this image into the presence or absence of specific beta-lactamase genes |
|
|